What is the best analysis of the gut microbiota?
Nahibu's gut microbiota study is conducted using metagenomics. This is a technique for sequencing DNA from complex samples such as soil, air, or stool samples. In metagenomics, organisms (bacteria in the case of Nahibu's microbiota analysis) are studied in their environment, without going through a culture stage, as in traditional microbiology.
The development of this technique has led to the discovery of many bacteria that cannot be cultured in the laboratory, thereby deepening our knowledge of the gut microbiota. To learn more about the microbiota, this still little-known organ, click here.
What are the methods used to study the microbiota?
There are two main methods for studying the gut microbiota: shotgun metagenomics and 16S RNA sequencing. Why not simply culture the bacteria as in a traditional microbiology lab and observe what grows to study our gut flora? Many gut bacteria cannot live in our environment; they are accustomed to growing without oxygen. What's more, we have around 100,000 billion bacteria in our bodies—imagine the surface area needed to cultivate all these bacteria! There are also hundreds of species of bacteria, each requiring very specific conditions. This is why, in order to study the bacteria in the gut microbiota, we analyze their DNA so that we can identify which bacteria they belong to and then, after analyzing all the DNA in a sample, list the bacteria present.
Targeted fecal or respiratory tests do not study the microbiota as a whole.
Other techniques only measure certain parameters in the stool but do not sequence the genes of the microbiota. These techniques evaluate specific markers such as calprotectin or other compounds present in the feces. They can also target specific bacteria that are cultured in microbiology laboratories to detect their presence. This does not require a sequencing step and does not provide an understanding of the total composition of the intestinal flora, as they only study specific markers or bacteria and not this organ as a whole.
There are also breath tests that measure the exhalation of one or more gases before and after eating a sugary snack. The aim is to assess the body's gas production, but these tests do not allow the bacterial composition of the gut microbiota to be studied, as gut bacteria are present in the stomach and intestine and not in exhaled gases.
Despite the name microbiota test given to certain targeted fecal tests or breath tests, they do not provide a complete analysis of the composition of the gut microbiota, which is made up of hundreds of different species of bacteria. Only metagenomics provides a complete and accurate description of the bacterial composition of our flora.
What is 16S RNA?
16S RNA sequencing studies a gene present in all bacteria, the 16S RNA gene. This gene differs between bacterial genera; this technique therefore makes it possible to identify the bacterial composition of a sample by describing the main genera that compose it.
What is metagenomics?
Metagenomics, or environmental genomics, is a method for studying the overall genetic content of a sample in a complex environment, such as the intestine. It allows all the genes present in the sample to be analyzed.
Why choose shotgun metagenomic analysis?
Shotgun metagenomics, the technique chosen by Nahibu, allows the DNA of all bacteria present in the sample to be studied without bias. Click here to learn about the steps involved in Nahibu's analysis.
When the results are delivered, users receive key indicators such as the diversity or balance of the gut microbiota, a complete and accurate map, and the functional potential of the microbiota. Functional potential explains the role that the detected bacteria play in our bodies. However, we refer to potential because metagenomics analyzes the DNA of bacteria, and not all DNA is constantly expressed; some remains silent and is therefore not active.
Metagenomics targeting 16S RNA is not metagenomics in the strict sense, but metagenetics, as it does not analyze all the genes in the sample, but only a part of them. In fact, it targets only the 16S RNA gene of bacteria.
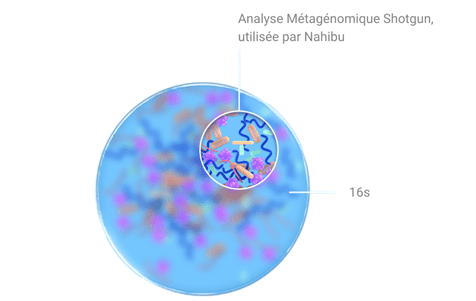
A more accurate description of bacteria
One of the advantages of shotgun metagenomics is that it provides a much more accurate description of the bacteria present in a sample.
Taxonomy is the practice of classifying organisms. It is used to name and classify bacteria. Shotgun metagenomics has better taxonomic resolution, meaning that it defines bacteria more precisely than other approaches. Shotgun metagenomics can characterize bacteria at the species or even strain level, while 16S only describes them at the genus level, and more rarely at the species level.
However, different strains of bacteria have different functions, so it is important to measure these subtleties.
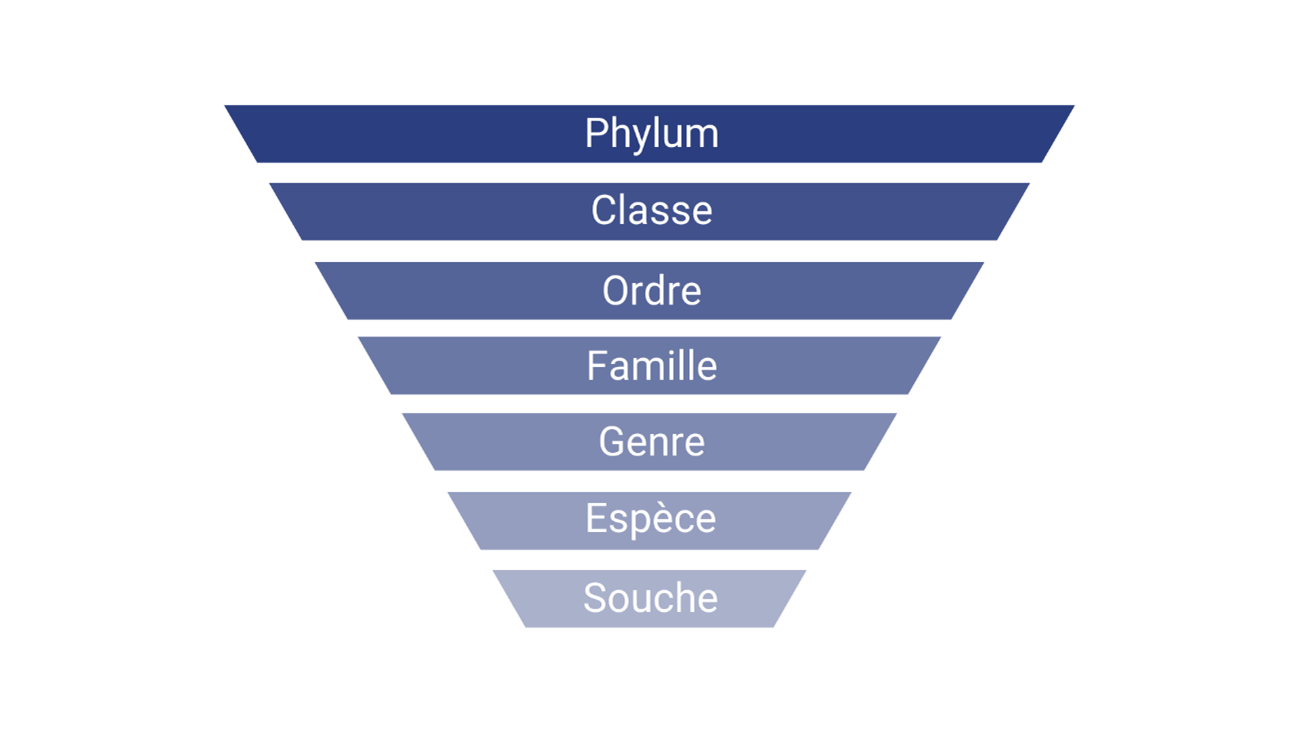
The potential role of the microbiota revealed by shotgun metagenomics
In addition, shotgun metagenomics makes it possible to describe the entire metagenome and thus explain the overall functioning of the microbiota. This allows us to extract the functional potential of the microbiota, which is impossible with an approach using 16S. Functional potential is the way in which the microbiota can hypothetically act on our metabolism and the functions of our body. Functional potential can be thought of as the role of each person in a household: cleaning, repairs, cooking, shopping, etc. Functional potential allows us to understand the role of each bacterium and which tasks an individual's microbiota will or will not be able to perform.
At Nahibu, we believe that providing you with a list of bacteria detected in a sample is not enough to enable you to understand the role your microbiota plays and identify its potential strengths and weaknesses. We have decided that microbiota analysis is part of a process of discovery and progress towards well-being. That's why we analyze your samples using shotgun metagenomics, a reliable, comprehensive, and unbiased approach that reveals the functional potential of your gut flora.
Nahibu's functional potential results are presented in categories with a score for each function. This allows you to clearly and accurately see which functions your microbiota has strong or weak potential for.
Next, how can you act and boost these functions? Foods or supplements are recommended to you on a personalized basis to improve your scores.
Shotgun metagenomics is a more expensive technique than 16S because it is comprehensive, generating files that take longer to analyze, but also providing much more complete results, a more accurate list of bacterial abundance, and an explanation of the functional potential of the microbiota, allowing you to understand its role in your body.
In conclusion, Nahibu's shotgun metagenomics analysis provides you with a comprehensive, accurate, easily understandable, and unbiased view of your gut microbiota to improve your well-being!

Discover more articles on the microbiota.
Understanding your gut microbiota analysis report
We explain how to interpret the Nahibu analysis report to better understand your gut microbiota.
The diversity of the gut microbiota: a key indicator of your health?
A high diversity of gut microbiota is often associated with the proper functioning of the body and helps to stay healthy.
Nahibu: steps for analyzing your stool sample
All stages of the analysis of your stool sample, up to the mapping of your microbiota.
Take care of your microbiota with Nahibu.